Publications
^ = undergraduate trainee, + = graduate trainee, ` = postdoc trainee, ® = visiting graduate trainee, #= high school student
@ = Peer-Reviewed, $ = Editorial Review, ~ = Preprint/Not reviewed
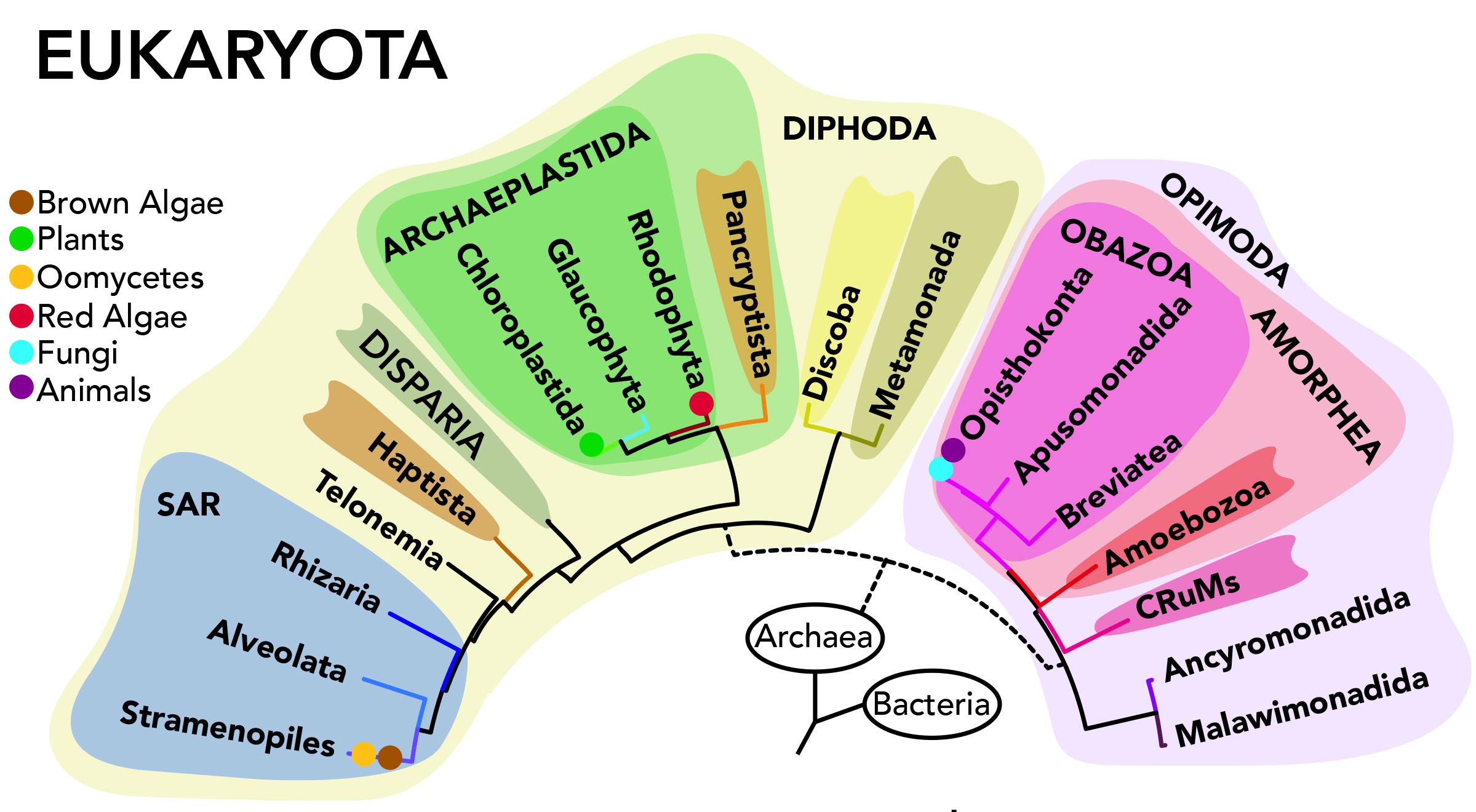
84. Valt M, Pánek T, Mirzoyan S, Tice AK`, Jones RE+, Dohnálek V, Dolezal P, Mikšátko J, Rotterová J, Hrubá P, Brown MW*, Čepička I*. Rare microbial relict sheds light on an ancient eukaryotic supergroup. Nature. https://doi.org/10.1038/s41586-025-09750-0 *Contributed equally – corresponding. @
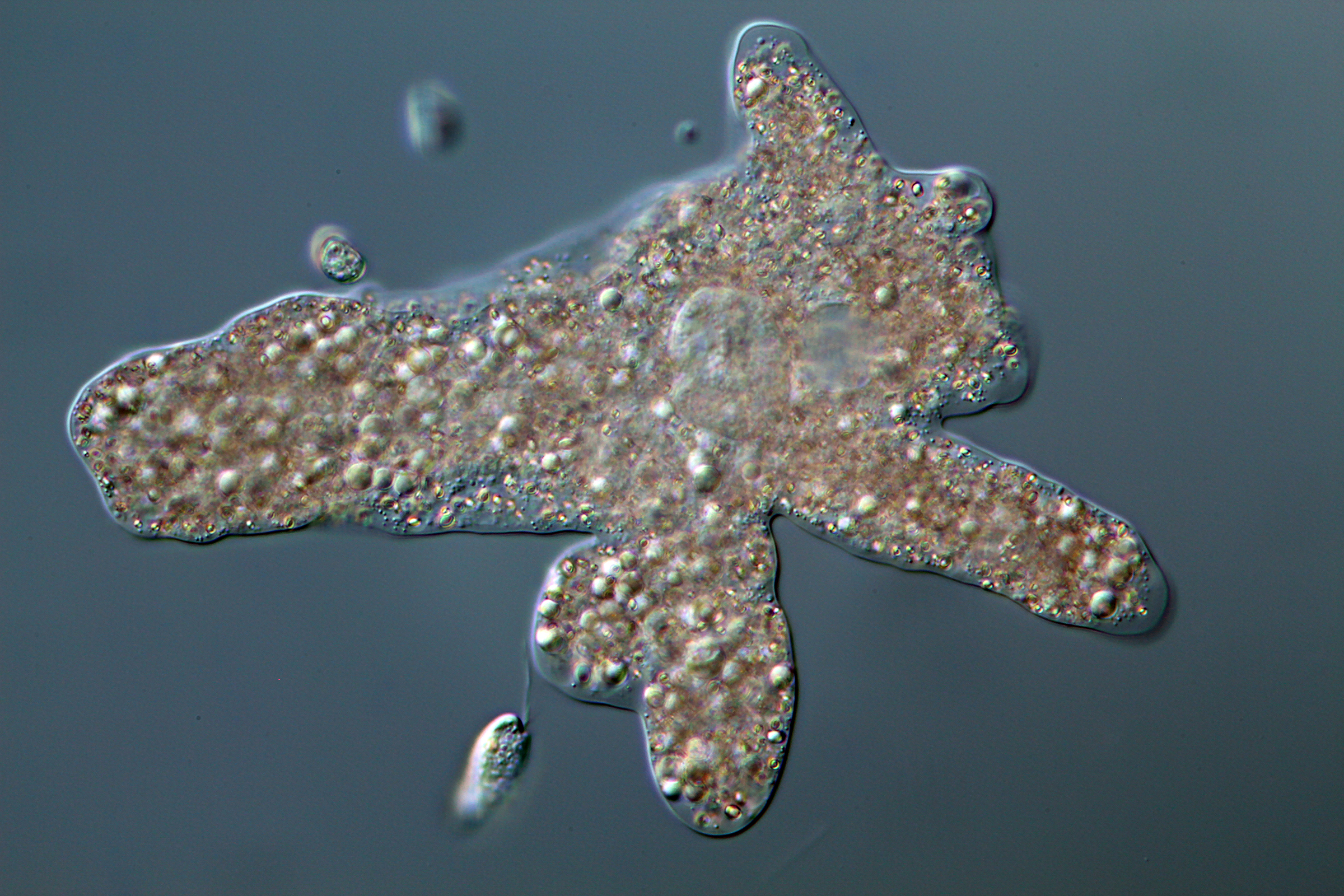
83. Jones RE+, Blandenier Q`, Kleitz-Singleton F+, Henderson TC+, Fry NW+, Banson I+, Nguyen J, Tice AK`, Brown MW. 2025. Piercing the veil: A novel amoebozoan (Janelia veilia n. gen. n. sp.) reveals deep clades within Discosea through phylogenomics. Protist. DOI: https://doi.org/10.1016/j.protis.2025.126103 @
82. Pánek T`, Tice AK`, Corre P, Hrubá P, Žihala D, Kamikawa R, Yazaki E, Shiratori T, Kume K, Hashimoto T, Ishida K, Hradilová M, Silberman JD, Roger AJ, Inagaki Y, Eliáš M, Brown MW, Čepička I 2025. An expanded phylogenomic analysis of Heterolobosea reveals the deep relationships, non-canonical genetic codes, and cryptic flagellate stages in the group. Molecular Phylogenetics and Evolution. DOI: https://doi.org/10.1016/j.ympev.2025.108289 @
81. Blandenier Q`, Gibson N^, Tice AK`, Jones RE+, Jones EP^, Baird RE, Zurweller BA, Brown MW. 2025. Full 18S metabarcoding of environmental samples of various substrates with Nanopore sequencing. Protocols.io. DOI: https://dx.doi.org/10.17504/protocols.io.3byl4qe48vo5/v1 ~
80. Tice AK`, Regis K, Shutt T, Spiegel FW, Brown MW*, Silberman JD*. 2024. Validating the genus Pocheina (Acrasidae, Heterolobosea, Excavata) leads to the recognition of three major lineages within Acrasidae. Journal of Eukaryotic Microbiology. bioRxiv. doi: https://doi.org/10.1101/2024.10.05.616810 * = contributed equally, co-senior @
79. Sheikh S, Fu CJ, Brown MW, Baldauf SL. 2024. The Acrasis kona genome and developmental transcriptomes reveal deep origins of eukaryotic multicellular pathways. Nature Communications. 15: 10197. DOI: https://doi.org/10.1038/s41467-024-54029-z @
78. Banos H, Wong TKF, Daneau J, Susko E, Minh BQ, Lanfear R, Brown MW, Eme L, Roger AJ. 2024. GTRpmix: A linked general-time reversible model for profile mixture models. Molecular Biology and Evolution. msae174, DOI: https://doi.org/10.1093/molbev/msae174. bioRxiv. 2024.03.29.587376. DOI: https://doi.org/10.1101/2024.03.29.587376 @
77. Porfirio-Sousa AL®*, Tice AK`*, Morais L, Ribeiro GM, Blandenier Q`, Dumack K, Eglit Y, Fry NW+, Souza MBGE, Henderson T+, Kleitz-Singleton F+, Singer D, Brown MW**, Lahr DJG**. 2024. Amoebozoan testate amoebae illuminate the diversity of heterotrophs and the complexity of ecosystems throughout geological time. Proceedings of the National Academy of Sciences, USA. 121 (30): e2319628121 DOI: https://www.pnas.org/doi/10.1073/pnas.2319628121. * &** = contributed equally (co-first, co-senior) @
76. Becker BM, Banson I+, Walker JM, Deshwal A, Brown MW, Silberman JD. 2024. Isolation of Naegleria lustrarea n. sp. (Excavata, Discoba, Heterolobosea) from Feces of Ambystoma annulatum (Ringed Salamander) in Northwest Arkansas. Journal of Eukaryotic Microbiology. DOI: https://doi.org/10.1111/jeu.13031 @
75. Henderson T+, Garcia-Gimeno L^, Beasley CE^, Fry NW+, Bess J^, Brown MW. 2024. High above the rest: Standing behaviors in the amoebae of Sappinia and Thecamoeba. European Journal of Protistology. DOI: https://doi.org/10.1016/j.ejop.2024.126082 @
74. Fry NW+, Jones RE+, Blandenier Q`, Tice AK`, Porfirio-Sousa AL®, Kleitz-Singleton F+, Henderson T+, Brown MW. 2024. Molecular phylogenetic analyses support the validity of Ceratiomyxa porioides (Amoebozoa, Eumycetozoa) at species level. European Journal of Protistology. DOI: https://doi.org/10.1016/j.ejop.2024.126083 @
73. Jones RE+, Tice AK`, Eliáš M, Eme L, Kolísko M, Nenarokov S, Pánek T, Rokas A, Salomaki E, Strassert JFH, Shen XX, Žihala D, Brown MW. 2024. Create, Analyze, and Visualize Phylogenomic Datasets Using PhyloFisher. Current Protocols. 4(1):e969 https://doi.org/10.1002/cpz1.969 @
72. Fry NW+, Schuler GA+, Jones RE+, Kooienga PG^, Jira V#, Shepherd M^, Tice AK`, Brown MW. 2024. Living in the cracks: two novel genera of Variosea (Amoebozoa) discovered on an urban sidewalk. Journal of Eukaryotic Microbiology. DOI: https://doi.org/10.1111/jeu.13020 @
71. Jones RE+, Jones EP^, Tice AK`, Brown MW. 2023. A PhyloFisher utility for nucleotide-based phylogenomic matrix construction; nucl_matrix_constructor.py. bioRxiv. 2023.11.30.569490. DOI: https://doi.org/10.1101/2023.11.30.569490 ~
70. Porfirio-Sousa AL@*, Tice AK`*, Morais L, Ribeiro GM, Blandenier Q, Dumack K, Eglit Y, Fry NW+, Souza MBGE, Henderson T+, Kleitz-Singleton F+, Singer D, Brown MW**, Lahr DJG. 2023**. Amoebozoan testate amoebae illuminate the diversity of heterotrophs and the complexity of ecosystems throughout geological time. bioRxiv 2023.11.08.566222; DOI: https://doi.org/10.1101/2023.11.08.566222 ~
69. Sheikh S, Fu CJ, Brown MW, Baldauf SL. 2023. Deep origins of eukaryotic multicellularity revealed by the Acrasis kona genome and developmental transcriptomes, Research Square {preprint} DOI: https://doi.org/10.21203/rs.3.rs-2587723/v1 ~
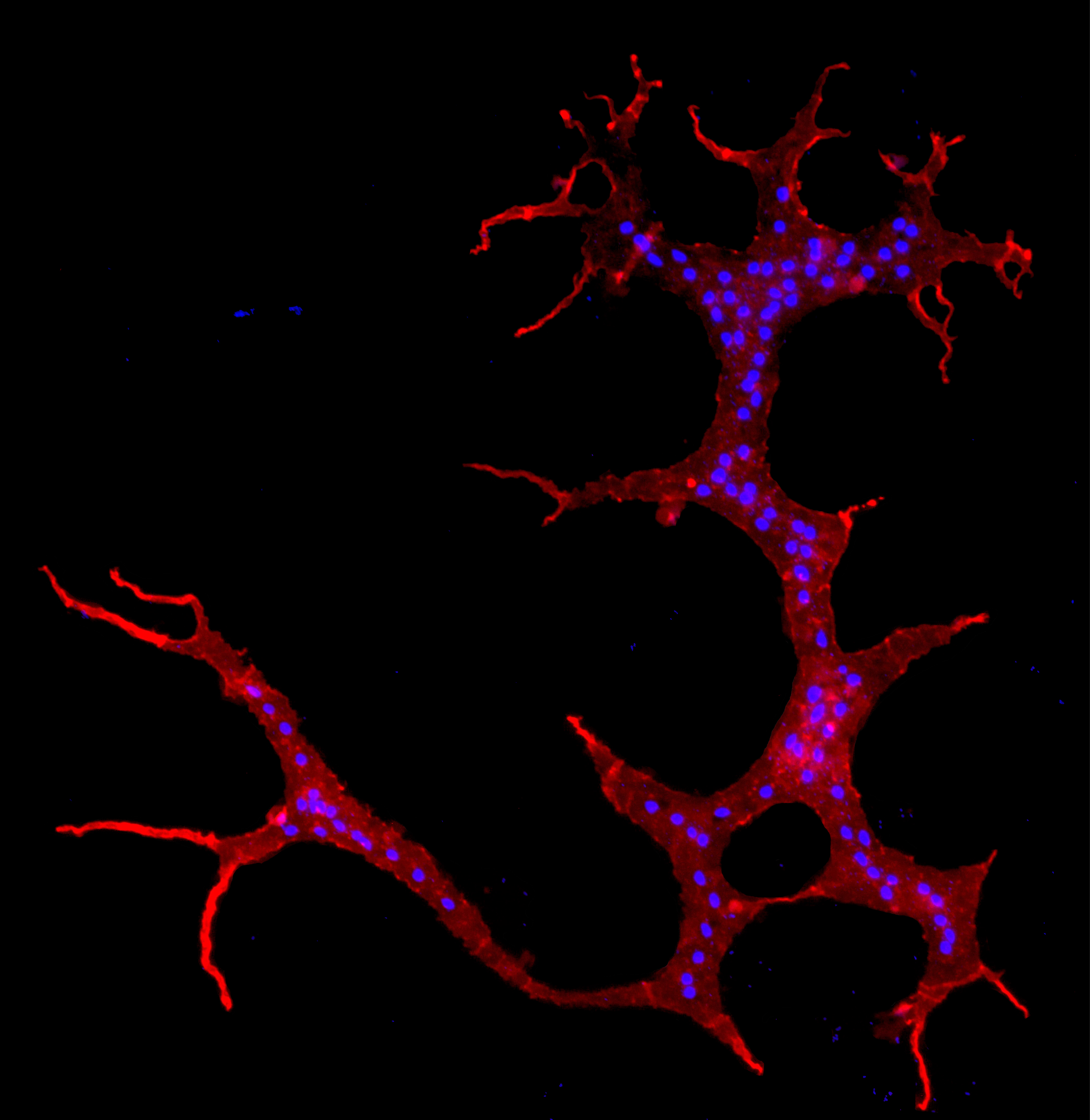
68. Porfirio-Sousa AL®, Henderson TC+, Brown MW. Effective and efficient cytoskeleton (actin and microtubules) fluorescence staining of adherent eukaryotic cells V.2. Protocols.io DOI: https://dx.doi.org/10.17504/protocols.io.kxygxeerzv8j/v2 ~
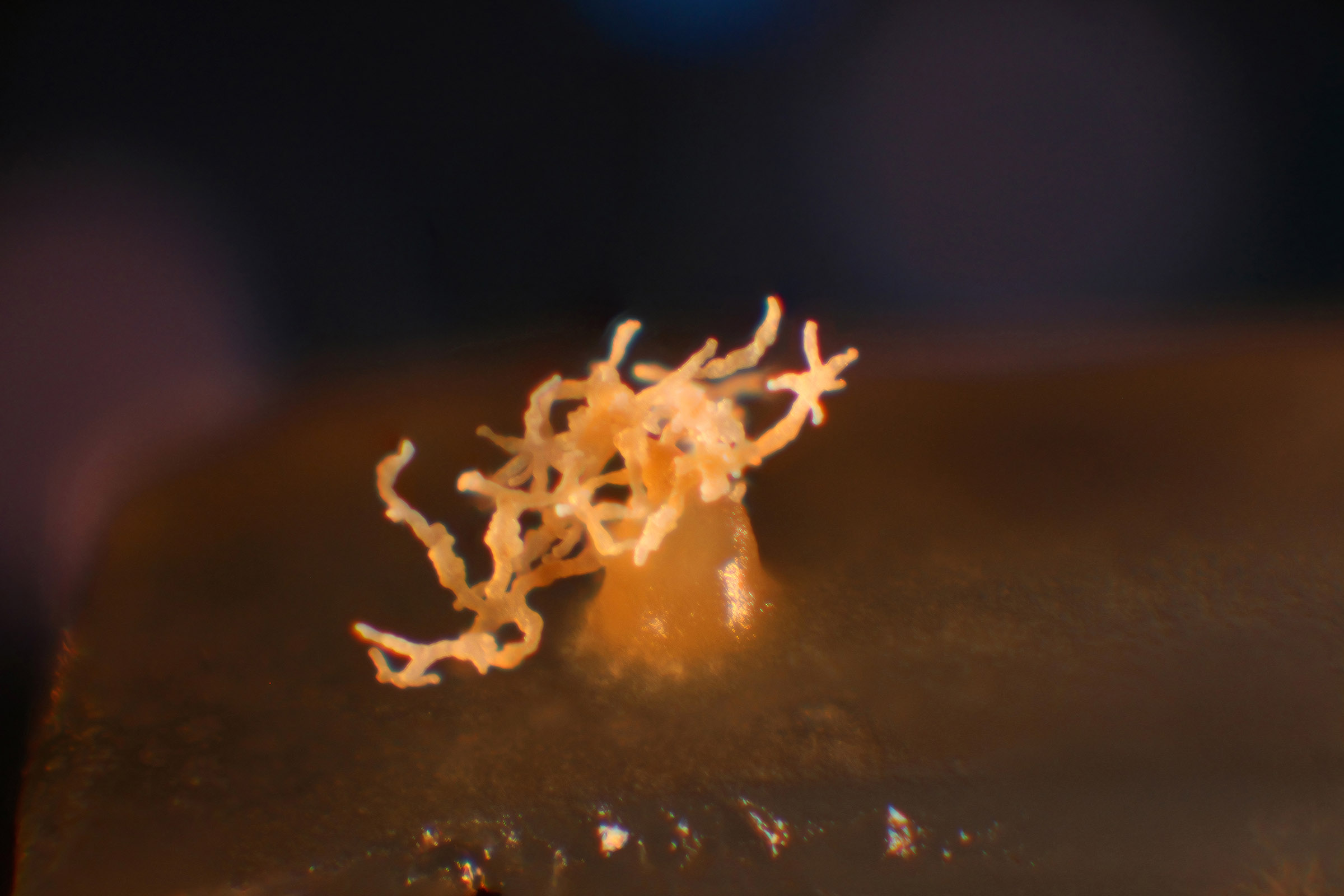
67. Tice AK`, Spiegel FW, Brown MW. 2023. Phylogenetic placement of the protosteloid amoeba Microglomus paxillus identifies another case of sporocarpic fruiting in Discosea (Amoebozoa). Journal of Eukaryotic Microbiology 24:e12971. DOI: https://doi.org/10.1111/jeu.12971@
66. Azuma T, Pánek T`, Tice AK`, Kayama M, Kobayashi M, Miyashita H, Suzaki T, Yabuki A, Brown MW*, Kamikawa R*. 2022. The enigmatic stramenopile Actinophrys sol sheds light on early steps for plastid organellogenesis in Ochrophyta. Molecular Biology and Evolution. 39(4):msac065. DOI: https://doi.org/10.1093/molbev/msac065 @ *= corresponding authors
65. Tice AK`, Brown MW. 2022. Multicellularity: Amoebae follow the leader to food. Current Biology. 32(9):R418-R420 DOI: https://doi.org/10.1016/j.cub.2022.03.067 $
64. Tice AK`, Žihala D®, Pánek T`, Jones RE+, Salomaki E, Nenarokov S, Burki F, Eliáš M, Eme L, Roger AJ, Rokas A, Shen X, Strassert JFH, Kolísko M, Brown MW. 2021. PhyloFisher: A phylogenomic package for resolving eukaryotic relationships. PLoS Biology. 19(8): e3001365. @
63. Rodriguez-Caro F, Fenner J, Bhardwaj S, Cole J, Benson JC, Colombara AM, Papa R, Brown MW, Martin A, Range RC, Counterman BA. 2021. Novel doublesex duplication associated with sexually dimorphic development of dogface butterfly wings. Molecular Biology and Evolution. 38(11):5021-5033. DOI: https://doi.org/10.1093/molbev/msab228 @
62. Brown MW. 2021. Effective and efficient cytoskeleton (actin and microtubules) fluorescence staining of adherent eukaryotic cells. Protocols.io. DOI: http://dx.doi.org/10.17504/protocols.io.8wkhxcw ~
61. Kang S+*, Tice AK`*, Stairs CW, Lahr DJG, Jones RE+, Brown MW. 2021. The integrin-mediated adhesive complex in the ancestor of animals, fungi, and amoebae. Current Biology. 31: 3073-3085. @ (*=contributed equally)
60. Porfírio-Sousa AL®, Tice AK+, Brown MW, Lahr DJG. 2021. Phylogenetic reconstruction and evolution of the Rab GTPase gene family in Amoebozoa. Small GTPases. DOI: 10.1080/21541248.2021.1903794 @
59. Salomaki E, Eme L, Brown MW, Kolisko M. 2020. Releasing uncurated datasets is essential for reproducible phylogenomics. Nature Ecology and Evolution. 4: 1435–1437. $
58. Lawal HM, Schilde C, Kin K, Brown MW, James J, Prescott AR, Schaap P. 2020. Cold climate adaptation is a plausible cause for evolution of multicellular sporulation in Dictyostelia. Scientific Reports. 10: 8797. [@
57. Onsbring H*, Tice AK+*, Barton BT, Brown MW”, Ettema TJG”. 2020. An efficient single-cell transcriptomics workflow for microbial eukaryotes benchmarked on Giardia intestinalis cells. BMC Genomics. 21: 448. @ *&”=Contributed equally (first and last).
56. Brown MW, Tice AK+. 2020. A genetic toolbox for marine protists. Nature Methods. 17(5): 469-470. $
55. Yubuki N, Lopez-Garcia P, Reboul G, Brown MW, Pollet N, Moreira D. 2020. Ancient Adaptive Lateral Gene Transfers in the Symbiotic Opalina - Blastocystis Stramenopile Lineage. Molecular Biology and Evolution. 37: 651–659. @
54. Burki F, Roger AJ, Brown MW, Simpson AGB. 2020. The new tree of eukaryotes. Trends in Ecology and Evolution. 35:43-55. @
53. Lahr DJG, Kosakyana A, Lara E, Mitchell EAD, Morais L, Porfirio-Sousa AL, Ribeiro GM, Tice AK+, Pánek T`, Kang S+, Brown MW. 2019. Phylogenomics and ancestral morphological reconstruction of testate amoebae demonstrate high diversity of microbial eukaryotes in the Neoproterozoic. Current Biology. 29(6): 911-1003.e3. @
52. Brown MW, Lahr DJG. 2019. ‘Micro snails’ we scraped from sidewalk cracks help unlock details of ancient earth’s biological evolution. The Conversation. 112362. https://theconversation.com/micro-snails-we-scraped-from-sidewalk-cracks-help-unlock-details-of-ancient-earths-biological-evolution-112362. $ popular science article
51. Fiore-Donno AM, Tice AK+, Brown MW. 2019. A Non-Flagellated Member of the Myxogastria and Expansion of the Echinosteliida. Journal of Eukaryotic Microbiology. 66: 538-544. @
50. Schuler GA+, Brown MW. 2019. Description of Armaparvus languidus n. gen. n. sp. confirms ultrastructural unity of Cutosea (Amoebozoa, Evosea). Journal of Eukaryotic Microbiology. 66: 158-166. @
49. Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Simpson AGB, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q. 2019. Revisions to the classification, nomenclature, and diversity of eukaryotes. Journal of Eukaryotic Microbiology. 66: 4-116. @
48. del Campo J, Kolisko M, Boscaro V, Santoferrara L, Massana R, Guillou L, Simpson AGB, Berney C, de Vargas C, Kaye J, Bender S, Brown MW, Keeling P, Wegener Parfrey L. 2018. EukRef: phylogenetic curation of ribosomal RNA to enhance understanding of eukaryotic diversity and distribution. PLoS Biology. 16(9): e2005849. @
47. Hofstatter PG, Brown MW, Lahr DJG. 2018. Comparative genomics supports sex and meiosis in diverse Amoebozoa. Genome Biology and Evolution. 10: 3118-3128. @
46. Schuler GA+, Tice AK+, Pearce RA, Foreman E^, Stone J, Gammill S, Wilson JD, Reading C, Silberman JD, Brown MW. 2018. Phylogeny and classification of novel diversity in Sainouroidea (Cercozoa, Rhizaria) sheds light on a highly diverse and divergent clade. Protist. 169: 853-874. @
45. Heiss AA, Kolisko M, Ekelund F, Brown MW, Roger AJ, Simpson AGB. 2018. Combined morphological and phylogenomic re-examination of malawimonads, a critical taxon for inferring the evolutionary history of eukaryotes. Royal Society Open Science. 5: 171707. @
44. Brown MW, Heiss AA, Kamikawa R, Inagaki Y, Yabuki A, Tice AK+, Shiratori T, Ishida K, Hashimoto T, Simpson AGB, Roger AJ. 2018. Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biology and Evolution. 10:427-433. SUPPLEMENTAL FILE - Single gene alignments and trees (Brown_etal.2017.CRuMS.tgz) @
43. Leonard G, Labarre A, Milner D, Monier A, Soanes D, Wideman JG, Maguire F, Stevens S, Sain D, Grau-Bove X, Sebe-Pedros A, Stajich JE, Paszkiewicz K, Brown MW, Hall N, Wickstead B, Richards TA. 2018. Comparative genomic analysis of the ‘psuedofungus’ Hyphochytrium catenoides. Royal Society Open Biology. 8: 170184. @
42. Kang S+, Tice AK+, Spiegel FW, Silberman JD, Pánek T, Čepička I, Kostka M, Kosakyan A, Alcântara DM, Roger AJ, Shadwick LL, Smirnov A, Kudryavstev A, Lahr DJG, Brown MW. 2017. Between a pod and a hard test: the deep evolution of amoebae. Molecular Biology and Evolution. 34: 2258-2270. SUPPLEMENTAL FILE - Single gene alignments and trees (Kang_etal.2017.tgz) @
41. Geisen S, Mitchell EAD, Wilkinson DM, Adl S, Bonkowski M, Brown MW, Fiore-Donno AM, Heger TJ, Jassey VEJ, Krashevska V, Lahr DJG, Marcisz K, Mulot M, Payne R, Singer D, Anderson OR, Charman DJ, Ekelund F, Griffiths BS, Rønn R, Smirnov A, Bass D, Belbahri L, Berney C, Blandenier Q, Chatzinotas A, Clarholm M, Dunthorn M, Feest A, Fernandez-Parra LD, Foissner W, Fournier B, Gentekaki E, Hajek M, Helder J, Jousset A, Koller R, Kumar S, La Terza A, Lamentowicz M, Mazei Y, Santos SS, Seppey CVW, Spiegel FW, Walochnik J, Winding A, Lara E. 2017. Soil protistology rebooted: 30 fundamental questions to start with. Soil Biology and Biochemistry. 111: 94–103. @
40. Spiegel FW, Shadwick L, Brown MW, Nderitu G, Aguliar M, Shadwick JDL. 2017. Protosteloid Amoebozoa (Protosteliids, Protosporangiida, Cavostellida, Schizoplasmodiida, Fractoviteliida, and sporocarpic members of Vanellida, Centramoebida, and Pellitida). In Archibald, Simpson, Slamovits (Eds.) Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.). Springer. DOI: 10.1007/978-3-319-32669-6_12-1 @
39. Tice AK+, Shadwick LL, Fiore-Donno AM, Geisen S, Kang S+, Schuler GA+, Spiegel FW, Wilkinson K, Bonkowski M, Dumack K, Lahr DJG, Voelcker E, Clauß S, Zhang J, Brown MW. 2016. Expansion of the molecular and morphological diversity of Acanthamoebidae (Centramoebida, Amoebozoa) and identification of a novel life cycle type within the group. Biology Direct. 11:69. @
38. Pánek T, Simpson AGB, Brown MW, Dyer BD. 2016. Heterolobosea. In Archibald, Simpson, Slamovits (Eds.) Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.). Springer. DOI: 10.1007/978-3-319-32669-6_10-1. @
37. Hofstatter P, Tice AK+, Kang S+, Brown MW*, Lahr DJG*. 2016. Evolution of bacterial recombinase A (recA) in eukaryotes explained by addition of genomic data of key microbial lineages. Proceedings of the Royal Society B: Biological Sciences. 283: 20161453. @ *Both last authors are corresponding.
36. Heiss AA, Brown MW, Simpson AGB. 2016. Apusomonadida. In Archibald, Simpson, Slamovits (Eds.) Handbook of the Protists (Second Edition of the Handbook of Protoctista by Margulis et al.). Springer. DOI: 10.1007/978-3-319-32669-6_15-1 @
35. Shadwick LL, Brown MW, Tice AK+, Spiegel FW. 2016. A new amoeba with protosteloid fruiting: Luapeleamoeba hula n. g. n. sp. Acta Protozoologica. 55(3): 123-134. @
34. Gawryluk RMR, Kamikawa R, Stairs CW, Silberman JD, Brown MW, Roger AJ. 2016. The earliest evolutionary stages of mitochondrial adaptation to low oxygen. Current Biology. 26(20): 2729–2738. @
33. Harding T, Brown MW, Simpson AGB, Roger AJ. 2016. Osmoadaptative strategy and its molecular signature in obligately halophilic heterotrophic protists. Genome Biology and Evolution. 8(7): 2241–2258. @
32. Hamann E, Gruber-Vodicka H, Kleiner M, Tegetmeyer HE, Riedel D, Littmann S, Chen J, Milucka J, Viehweger B, Becker KW, Dong X, Stairs CW, Hinrichs K, Brown MW, Roger AJ, Strous M. 2016. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature. 534: 254-258. @
31. Noguchi F, Tanifuji G, Brown MW, Fujikura K, Takishita K. 2016. Complex evolution of two types of cardiolipin synthase in the eukaryotic lineage stramenopiles. Molecular Phylogenetics and Evolution. 101: 133-141. @
30. Tice AK+, Brown MW, Altig R. 2016. Hyla chrysoscelis (Cope’s gray treefrog), H. avivoca (bird-voiced treefrog), and H. cinerea (green treefrog). Interactions of protozoans and algae with frog eggs and tadpoles. Herpetological Review. 47(3): 438-439. $
29. Walthall AC^, Tice AK+, Brown MW. 2016. A new species of Flamella (Amoebozoa, Variosea, Gracilipodida) isolated from a freshwater pool in southern Mississippi, USA. Acta Protozoologica. 55(2): 111-117. @
28. Tice AK+, Silberman JD, Walthall AC^, Le KND, Spiegel FW, Brown MW. 2016. Sorodiplophrys stercorea: another novel lineage of sorocarpic multicellularity. Journal of Eukaryotic Microbiology. 63(5): 623-628. @
27. Bass D, Silberman JD, Brown MW, Pearce RA, Tice AK+, Jousset A, Geisen S, Hartikainen H. 2016. Coprophilic amoebae and flagellates, including Guttulinopsis, Rosculus and Helkesimastix, characterise a divergent and diverse rhizarian radiation and contribute to a large diversity of faecal-associated protists. Environmental Microbiology. 18(5): 1604-1619. @
26. Pánek T, Zadrobílková E, Walker G, Brown MW, Gentekaki E, Hroudová M, Kang S+, Roger AJ, Tice AK+, Vlček C, Čepička I. 2016. First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group. Molecular Phylogenetics and Evolution. 98: 41-51. @
25. Brown LR, Gunnell SM, Cassella A, Keller LE, Scherkenbach LA, Mann B, Brown MW, Rosch JW, Tuomanen EI, Thornton JA. 2016. AdcAII of Streptococcus pneumoniae affects pneumococcal invasiveness. PLoS ONE. 11(1): e0146785. @
24. Sharpe SC, Eme L, Brown MW, Roger AJ. 2015. Timing the origins of multicellular eukaryotes through phylogenomics and relaxed molecular clock analyses. In Nedelcu, Ruiz-Trillo (Eds.) Evolutionary transitions to multicellular life: Principles and mechanisms. Springer, New York. pp. 3-29. ISBN: 978-94-017-9641-5. DOI: 10.1007/978-94-017-9642-2_1. @
23. Maguire F, Henriquez F, Leonard G, Dacks JB, Brown MW, Richards TA. 2014. Complex patterns of gene fusion and fission in the eukaryotic folate biosynthesis pathway. Genome Biology and Evolution. 6(10): 2709-2720. @
22. Watson PM^, Sorrell SC^, Brown MW. 2014. Ptolemeba n. gen., a novel genus of hartmannellid amoebae (Tubulinea, Amoebozoa); with an emphasis on the taxonomy of Saccamoeba. Journal of Eukaryotic Microbiology. 61(6): 611-619. @
21. Eme L, Sharpe SC, Brown MW, Roger AJ. 2014. On the age of eukaryotes: Evaluating evidence from fossils and molecular clocks. Cold Spring Harbor Perspectives in Biology. 6: a016139. Online publication, but also printed in Keeling, Koonin (Eds.) The Origin and Evolution of Eukaryotes, a subject collection from Cold Spring Harbor Perspectives in Biology. pp. 165-180. @
20. Stairs CW, Eme L, Brown MW, Mutsaers C, Susko E, Dellaire G, Soanes DM, van der Giezen M, Roger AJ. 2014. A SUF Fe-S cluster biogenesis system in the mitochondrion-related organelles of the microaerophilic protist Pygsuia. Current Biology. 24: 1176-1186. @
19. Kudryavtsev A, Brown MW, Tice AK, Spiegel FW, Pawlowski J, Anderson OR. 2014. A revision of the order Pellitida Smirnov et al., 2011 (Amoebozoa, Discosea) based on ultrastructural and molecular evidence, with description of Endostelium crystalliferum n. sp. Protist. 165(2): 208-229. @
18. Tanifuji G, Onodera N, Brown MW, Curtis BA, Roger AJ, Wong GK, Melkonian M, Archibald JM. 2014. Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics. 15: 374. @
17. Suga H, Torruella G, Burger G, Brown MW, Ruiz-Trillo I. 2014. Earliest holozoan expansion of phosphotyrosine signaling. Molecular Biology and Evolution. 31(3): 517-528. @
16. Kamikawa R, Kolisko M, Nishimura Y, Yabuki A, Brown MW, Ishikawa SA, Ishida K, Simpson AGB, Roger AJ, Hashimoto T, Inagaki Y. 2014. Gene-content evolution in discobid mitochondria deduced from the phylogenetic position and complete mitochondrial genome of Tsukubamonas globosa. Genome Biology and Evolution. 6(2): 306–315. @
15. Brown MW, Sharpe S, Silberman JD, Heiss A, Lang BF, Simpson AGB, Roger AJ. 2013 Phylogenomics demonstrates that breviate flagellates are related to opisthokonts. Proceedings of the Royal Society B: Biological Sciences. 280: 20131755. SUPPLEMENTAL FILE - Single gene alignments and trees (Brown_etal.2013.SingleGenes.tgz) @
14. Brown MW*, Silberman JD. 2013. The Non-Dictyostelid Sorocarpic Amoebae. In Romeralo, Escalante, Baldauf (Eds.) Dictyostelids - Evolution, Genomics and Cell Biology. Springer, Heidelberg Germany. pp. 219-242. ISBN: 978-3-642-38487-5. DOI: 10.1007/978-3-642-38487-5_12. *Senior & Corresponding Author. $
13. Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sánchez-Pons N, Torruella G, Derelle R, Manning G, Lang BF, Russ C, Haas BJ, Roger AJ, Nusbaum C, Ruiz-Trillo I. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nature Communications. 4: 2325. @
12. Kamikawa R, Brown MW, Nishimura Y, Sako Y, Heiss AA, Yubuki N, Gawryluk R, Simpson AGB, Roger AJ, Hashimoto T, Inagaki Y. 2013. Parallel re-modeling of EF-1α function: divergent EF-1α genes co-occur with EFL genes in diverse distantly related eukaryotes. BMC Evolutionary Biology. 13: 131. @
11. Harding T, Brown MW, Plotnikov A, Selivanova E, Park JS, Gunderson JH, Baumgartner M, Silberman JD, Roger AJ, Simpson AGB. 2013. Amoeba stages in the deepest branching heteroloboseans, including Pharyngomonas: evolutionary and systematic implications. Protist. 164(2): 272-286. @
10. Adl SM, Simpson AGB, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Legall L, Lynn DH, McManus H, Mitchell EAD, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick LL, Schoch C, Smirnov A, Spiegel FW. 2012. The revised classification of eukaryotes. Journal of Eukaryotic Microbiology. 59(5): 429–514. @
9. Schnittler M, Novozhilov YK, Romeralo M, Brown MW, Fiore-Donno AM. 2012. Fruit body-forming protists: Myxomycetes and Myxomycete-like organisms Acrasia, Eumycetozoa. In Frey (Ed.) Engler’s Syllabus of Plant Families, 13th ed. Part 1: Blue-green Algae, Myxomycetes and Myxomycete-like Organisms, Phytoparasitic protists, Heterotrophic Heterokontobionta and Fungi. Borntraeger Science Publishers, Berlin. pp. 40-88. ISBN 978-3-443-01061-4. DOI: 10.13140/2.1.4668.9284. $
8. Brown MW, Kolisko M, Silberman JD, Roger AJ. 2012. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Current Biology. 22(12): 1123-1127. @
7. Brown MW, Silberman JD, Spiegel FW. 2012. A contemporary evaluation of the acrasids (Acrasidae, Heterolobosea, Excavata). European Journal of Protistology. 48(2): 103-123. Invited manuscript. @
6. Brown MW, Silberman JD, Spiegel FW. 2011. “Slime molds” among the Tubulinea (Amoebozoa): Molecular systematics and taxonomy of Copromyxa. Protist. 162: 277-287. @
5. Brown MW, Silberman JD, Spiegel FW. 2010. A morphologically simple species of Acrasis (Heterolobosea, Excavata), Acrasis helenhemmesae n. sp. Journal of Eukaryotic Microbiology. 57(4): 346-353. @
4. Brown MW, Spiegel FW, Silberman JD. 2009. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Molecular Biology and Evolution. 26(12): 2699-2709. @
3. Shadwick LL, Spiegel FW, Shadwick JDL, Brown MW, Silberman JD. 2009. Eumycetozoa = Amoebozoa?: SSUrDNA Phylogeny of Protosteloid Slime Molds and its Significance for the Supergroup Amoebozoa. PLoS ONE. 4(8): e6754. @
2. Spiegel FW, Shadwick JD, Lindley LA, Brown MW, Nderitu G. 2007. A Beginner’s Guide to Identifying the Protostelids. http://slimemold.uark.edu/pdfs/Handbook1_3rd.pdf ~
1. Brown MW, Spiegel FW, Silberman JD. 2007. Amoeba at Attention: Phylogenetic Affinity of Sappinia pedata. Journal of Eukaryotic Microbiology. 54(6): 511-519. @